Multifidelity Modelling with Autoregressive Model#
This tutorial demonstrates the usage of the MultifidelityAutoregressive model for fitting multifidelity data. This is an implementation of the AR1 model initially described in [KOHagan00].
[1]:
import tensorflow as tf
import numpy as np
import matplotlib.pyplot as plt
np.random.seed(1793)
tf.random.set_seed(1793)
Describe the problem#
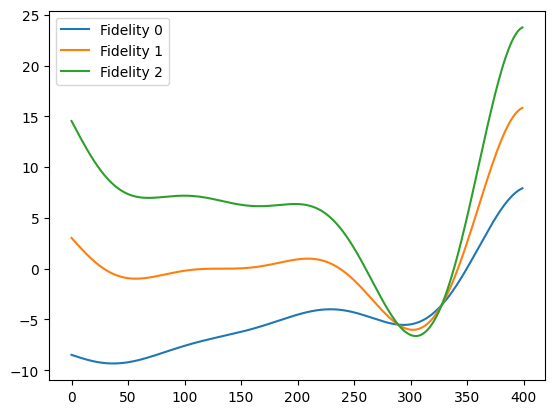
In this tutorial we will consider the scenario where we have a simulator that can be run at three fidelities, with the ability to get cheap but coarse results at the lowest fidelity, more expensive but more refined results at a middle fidelity and very accurate but very expensive results at the highest fidelity.
We define the true functions for the fidelities as:
Note that noise is optionally added to any observations in all but the lowest fidelity. There are a few modelling assumptions: 1. The lowest fidelity is noise-free 2. The data is cascading, e.g any point that has an observation at a high fidelity also has one at the lower fidelities.
[2]:
# Define the multifidelity simulator
def simulator(x_input, fidelity, add_noise=False):
f = 0.5 * ((6.0 * x_input - 2.0) ** 2) * tf.math.sin(
12.0 * x_input - 4.0
) + 10.0 * (x_input - 1.0)
f = f + fidelity * (f - 20.0 * (x_input - 1.0))
if add_noise:
noise = tf.random.normal(f.shape, stddev=1e-1, dtype=f.dtype)
else:
noise = 0
f = tf.where(fidelity > 0, f + noise, f)
return f
# Plot the fidelities
x = np.linspace(0, 1, 400)
y0 = simulator(x, 0)
y1 = simulator(x, 1)
y2 = simulator(x, 2)
plt.plot(y0, label="Fidelity 0")
plt.plot(y1, label="Fidelity 1")
plt.plot(y2, label="Fidelity 2")
plt.legend()
plt.show()
Trieste handles fidelities by adding an extra column to the data containing the fidelity information of the query point. The function check_and_extract_fidelity_query_points will check that the fidelity column is valid, and if so, will separate the query points and the fidelity information.
[3]:
from trieste.data import Dataset, check_and_extract_fidelity_query_points
# Create an observer class to deal with multifidelity input query points
class Observer:
def __init__(self):
self.simulator = simulator
def __call__(self, x, add_noise=True):
# Extract raw input and fidelity columns
x_input, x_fidelity = check_and_extract_fidelity_query_points(x)
# note: this assumes that my_simulator broadcasts, i.e. accept matrix inputs.
# If not you need to replace this by a for loop over all rows of "input"
observations = self.simulator(x_input, x_fidelity, add_noise)
return Dataset(query_points=x, observations=observations)
# Instantiate the observer
observer = Observer()
/opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages/gpflow/experimental/utils.py:43: UserWarning: You're calling gpflow.experimental.check_shapes.decorator.check_shapes which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
f"You're calling {name} which is considered *experimental*."
/opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages/gpflow/experimental/utils.py:43: UserWarning: You're calling gpflow.experimental.check_shapes.inheritance.inherit_check_shapes which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
f"You're calling {name} which is considered *experimental*."
Now we can define the other parameters of our problem, such as the input dimension, search space and number of fidelities.
[4]:
from trieste.space import Box
input_dim = 1
n_fidelities = 3
lb = np.zeros(input_dim)
ub = np.ones(input_dim)
input_search_space = Box(lb, ub)
Create initial dataset#
[5]:
from trieste.data import add_fidelity_column
# Define sample sizes of low, mid and high fidelities
sample_sizes = [18, 12, 6]
xs = [tf.linspace(0, 1, sample_sizes[0])[:, None]]
# Take a subsample of each lower fidelity to sample at the next fidelity up
for fidelity in range(1, n_fidelities):
samples = tf.Variable(
np.random.choice(
xs[fidelity - 1][:, 0], size=sample_sizes[fidelity], replace=False
)
)[:, None]
xs.append(samples)
# Add fidelity columns to training data
initial_samples_list = [add_fidelity_column(x, i) for i, x in enumerate(xs)]
initial_sample = tf.concat(initial_samples_list, 0)
initial_data = observer(initial_sample, add_noise=True)
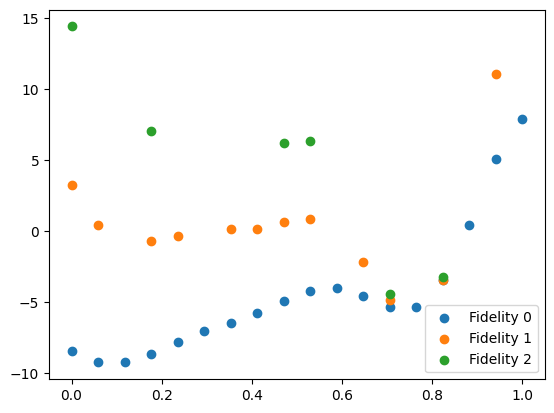
We can plot the initial data. We separate the dataset into individual fidelities using the split_dataset_by_fidelity function.
[6]:
from trieste.data import split_dataset_by_fidelity
data = split_dataset_by_fidelity(initial_data, num_fidelities=n_fidelities)
plt.scatter(data[0].query_points, data[0].observations, label="Fidelity 0")
plt.scatter(data[1].query_points, data[1].observations, label="Fidelity 1")
plt.scatter(data[2].query_points, data[2].observations, label="Fidelity 2")
plt.legend()
plt.show()
Now we can fit the MultifidelityAutoregressive model to this data. We use the build_multifidelity_autoregressive_models to create the sub-models required by the multifidelity model.
[7]:
from trieste.models.gpflow import (
MultifidelityAutoregressive,
build_multifidelity_autoregressive_models,
)
# Initialise model
multifidelity_model = MultifidelityAutoregressive(
build_multifidelity_autoregressive_models(
initial_data, n_fidelities, input_search_space
)
)
# Update and optimize model
multifidelity_model.update(initial_data)
multifidelity_model.optimize(initial_data)
/opt/hostedtoolcache/Python/3.7.15/x64/lib/python3.7/site-packages/gpflow/experimental/utils.py:43: UserWarning: You're calling gpflow.experimental.check_shapes.checker.ShapeChecker.__init__ which is considered *experimental*. Expect: breaking changes, poor documentation, and bugs.
f"You're calling {name} which is considered *experimental*."
Plot Results#
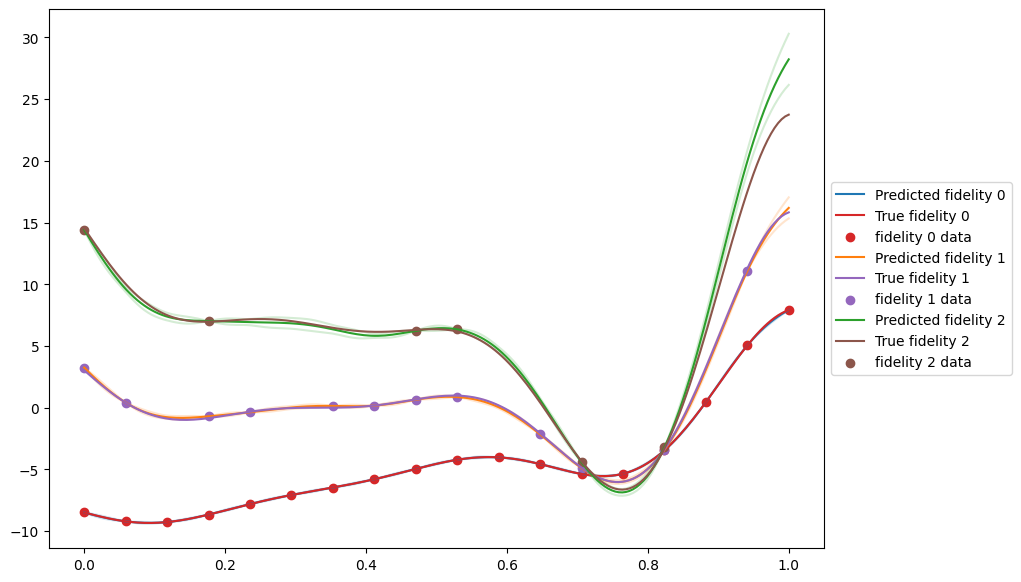
Now we can plot the results to have a look at the fit. The MultifidelityAutoregressive.predict method requires data with a fidelity column that specifies the fidelity for each data point to be predicted at. We use the add_fidelity_column function to add this.
[8]:
X = tf.linspace(0, 1, 200)[:, None]
X_list = [add_fidelity_column(X, i) for i in range(n_fidelities)]
predictions = [multifidelity_model.predict(x) for x in X_list]
fig, ax = plt.subplots(1, 1, figsize=(10, 7))
pred_colors = ["tab:blue", "tab:orange", "tab:green"]
gt_colors = ["tab:red", "tab:purple", "tab:brown"]
for fidelity, prediction in enumerate(predictions):
mean, var = prediction
ax.plot(
X,
mean,
label=f"Predicted fidelity {fidelity}",
color=pred_colors[fidelity],
)
ax.plot(
X,
mean + 1.96 * tf.math.sqrt(var),
alpha=0.2,
color=pred_colors[fidelity],
)
ax.plot(
X,
mean - 1.96 * tf.math.sqrt(var),
alpha=0.2,
color=pred_colors[fidelity],
)
ax.plot(
X,
observer(X_list[fidelity], add_noise=False).observations,
label=f"True fidelity {fidelity}",
color=gt_colors[fidelity],
)
ax.scatter(
data[fidelity].query_points,
data[fidelity].observations,
label=f"fidelity {fidelity} data",
color=gt_colors[fidelity],
)
ax.legend(loc="center left", bbox_to_anchor=(1, 0.5))
plt.show()
Comparison with naive model fit on high fidelity#
We can compare with a model that was fit just on the high fidelity data, and see the gains from using the low fidelity data.
[9]:
from trieste.models.gpflow import GaussianProcessRegression, build_gpr
from trieste.data import add_fidelity_column
# Get high fidleity data
hf_data = data[2]
# Fit simple gpr model to high fidelity data
gpr_model = GaussianProcessRegression(build_gpr(hf_data, input_search_space))
gpr_model.update(hf_data)
gpr_model.optimize(hf_data)
X = tf.linspace(0, 1, 200)[:, None]
# Turn X into high fidelity query points for the multifidelity model
X_for_multifid = add_fidelity_column(X, 2)
gpr_predictions = gpr_model.predict(X)
multifidelity_predictions = multifidelity_model.predict(X_for_multifid)
fig, ax = plt.subplots(1, 1, figsize=(10, 7))
"tab:blue", "tab:orange", "tab:green"
# Plot gpr results
mean, var = gpr_predictions
ax.plot(X, mean, label=f"GPR", color="tab:blue")
ax.plot(X, mean + 1.96 * tf.math.sqrt(var), alpha=0.2, color="tab:blue")
ax.plot(X, mean - 1.96 * tf.math.sqrt(var), alpha=0.2, color="tab:blue")
# Plot gpr results
mean, var = multifidelity_predictions
ax.plot(X, mean, label=f"MultifidelityAutoregressive", color="tab:orange")
ax.plot(X, mean + 1.96 * tf.math.sqrt(var), alpha=0.2, color="tab:orange")
ax.plot(X, mean - 1.96 * tf.math.sqrt(var), alpha=0.2, color="tab:orange")
# Plot true function
ax.plot(
X,
observer(X_for_multifid, add_noise=False).observations,
label=f"True function",
color="tab:green",
)
# Scatter the data
ax.scatter(
hf_data.query_points, hf_data.observations, label=f"Data", color="tab:green"
)
plt.legend()
plt.show()
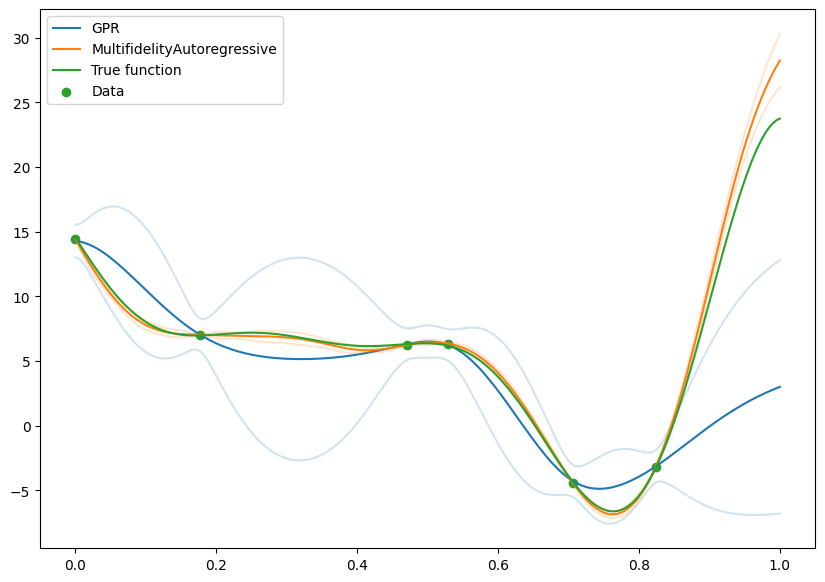
It’s clear that there is a large benefit to being able to make use of the low fidelity data, and this is particularly noticable in the greatly reduced confidence intervals.